2025 Benchmark: De-Identification Tools
Post Summary
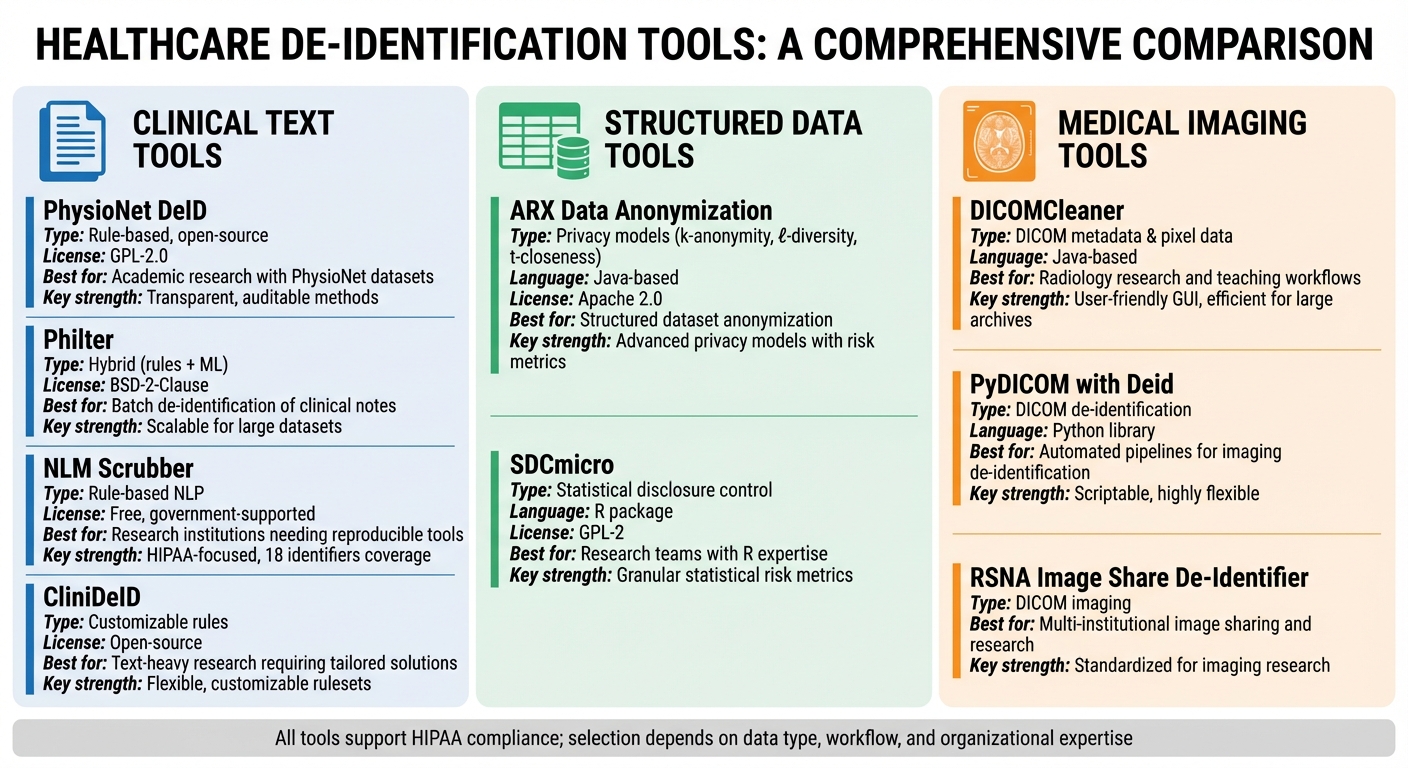
De-identification tools are essential for protecting sensitive healthcare data while enabling research, AI development, and compliance with regulations like HIPAA, GDPR, and CCPA. This benchmark evaluates nine leading solutions designed to handle clinical text, structured data, and medical imaging. Each tool's strengths, limitations, and ideal use cases are outlined to help healthcare organizations select the best fit for their needs.
Key Takeaways:
- Clinical Text Tools: PhysioNet DeID, Philter, NLM Scrubber, and CliniDeID focus on removing identifiers from narrative documents like EHR notes. These are best for research and compliance workflows.
- Structured Data Tools: ARX and SDCmicro specialize in anonymizing tabular datasets using advanced privacy models like k-anonymity and l-diversity.
- Imaging Tools: DICOMCleaner, PyDICOM with Deid, and RSNA Image Share De-Identifier address PHI in DICOM metadata and pixel data for medical imaging.
Quick Overview:
- PhysioNet DeID: Rule-based, open-source for free-text clinical notes.
- Philter: Hybrid of rules and machine learning for large-scale text de-identification.
- NLM Scrubber: Government-supported tool focused on HIPAA compliance for clinical text.
- CliniDeID: Customizable rules for unstructured text in research environments.
- ARX: Java-based tool for anonymizing structured datasets with privacy models.
- SDCmicro: R package for statistical disclosure control in microdata.
- DICOMCleaner: Java-based software for de-identifying DICOM metadata and pixel data.
- PyDICOM with Deid: Python library for flexible DICOM de-identification.
- RSNA Image Share De-Identifier: Designed for DICOM workflows in imaging research.
Quick Comparison
| Tool | Primary Data Type | Key Strengths | Best Fit |
|---|---|---|---|
| PhysioNet DeID | Clinical text | Open-source, rule-based | Academic research with PhysioNet datasets |
| Philter | Clinical text | Hybrid approach, scalable | Batch de-identification of clinical notes |
| NLM Scrubber | Clinical text | HIPAA-focused, transparent NLP methods | Research institutions needing reproducible tools |
| CliniDeID | Clinical text | Customizable, flexible rules | Text-heavy research requiring tailored solutions |
| ARX | Structured/tabular data | Advanced privacy models | Structured dataset anonymization |
| SDCmicro | Structured/tabular data | Statistical risk metrics | Research teams with expertise in R workflows |
| DICOMCleaner | DICOM imaging | User-friendly, efficient for large archives | Radiology research and teaching workflows |
| PyDICOM with Deid | DICOM imaging | Scriptable, highly flexible | Automated pipelines for imaging de-identification |
| RSNA Image Share De-Identifier | DICOM imaging | Standardized for imaging research | Multi-institutional image sharing and research |
Choose a tool based on your data type, compliance needs, and workflow requirements. Testing tools on real datasets ensures proper PHI removal and maintains data utility for secondary use.
2025 Healthcare De-Identification Tools Comparison: Clinical Text, Structured Data, and Medical Imaging Solutions
1. PhysioNet DeID
PhysioNet DeID is an open-source, rule-based de-identification tool created by MIT's Laboratory for Computational Physiology as part of the MIMIC project. Its main goal is to remove Protected Health Information (PHI) from free-text clinical notes, allowing researchers to access intensive care datasets while maintaining patient privacy [2][11]. The tool operates under a GPL-2.0 license, meaning it’s free to use and modify, as long as any derivative works also comply with GPL requirements [2].
Data Type Coverage
This tool is specifically designed to handle English-language, free-text clinical documents, such as discharge summaries, progress notes, and ICU records commonly found in the MIMIC datasets [2][11]. However, it does not process other types of data like DICOM metadata, HL7 segments, or structured EHR data. Since it focuses solely on unstructured text, additional tools are needed to handle images or structured database fields [4].
De-Identification Techniques
PhysioNet DeID relies on pattern-based recognition and dictionaries to detect PHI elements, including names, addresses, dates, phone numbers, and medical record numbers [4]. Instead of simply redacting this information, the tool replaces PHI with realistic surrogates, preserving the text's linguistic and temporal context [2][11]. While this approach ensures high precision for standard identifiers, it may struggle with rare, misspelled, or highly specific terms compared to modern machine-learning-based de-identification systems [3]. By maintaining the text's structure, it supports clinical NLP tasks and downstream data analysis.
Compliance and Governance
Ensuring compliance with data protection standards is a key feature of PhysioNet DeID. It aligns with HIPAA Safe Harbor guidelines by removing or transforming the 18 categories of identifiers found in unstructured text [2]. However, using the tool alone doesn’t guarantee full compliance. U.S. healthcare providers must validate the de-identified outputs, assess potential re-identification risks, and perform additional checks as necessary [4][5]. For a more comprehensive approach, organizations can integrate de-identification workflows into broader risk management platforms like Censinet RiskOps™. These platforms help track de-identification efforts, manage vendor risks, and strengthen cybersecurity measures for handling PHI [7].
Integration and Deployment
PhysioNet DeID is designed for on-premises deployment, functioning as a scriptable tool that integrates seamlessly into batch workflows [4]. Since all processing occurs locally, organizations retain full control over PHI within their infrastructure. This local processing approach simplifies compliance with HIPAA regulations by eliminating the need to transfer sensitive data to external APIs [7][9].
2. Philter
Philter is an open-source, hybrid de-identification tool created at the University of Pittsburgh to remove PHI (Protected Health Information) from clinical text on a large scale [2]. Released under a BSD-2-Clause license, it offers greater flexibility compared to GPL-licensed tools, making it easier for healthcare organizations and vendors to integrate into both research workflows and commercial applications [2]. By combining rule-based methods with machine-learning techniques, Philter strikes a balance between protecting privacy and maintaining data usability, bridging the gap between traditional rule-based systems and machine learning [2][11]. This hybrid strategy lays the foundation for effective data handling, as explored in later sections.
Data Type Coverage
Philter is tailored for clinical narratives, such as progress notes, discharge summaries, radiology impressions, pathology reports, and patient portal messages [2]. It identifies and removes direct identifiers like patient names, addresses, phone numbers, and medical record numbers. Users can also extend its functionality by configuring blacklists to include site-specific terms, like local clinic names or vendor identifiers [4]. However, it does not support structured data formats such as DICOM headers or HL7 segments. For organizations dealing with imaging or database fields, additional tools would be necessary [2].
De-Identification Techniques
Philter employs pattern-based detection (e.g., regular expressions) to identify structured identifiers like dates, phone numbers, ZIP codes, and URLs. It also uses dictionary lookups to capture common names, locations, and organizations [2]. For more nuanced PHI that patterns might miss, machine-learning algorithms come into play. Users can expand dictionaries and create custom blacklists to suit local documentation practices, such as adding regional payor names or internal system IDs [4]. Once identified, PHI can be masked using tokens like "[NAME]" or removed entirely. Some deployments even include surrogate generation in downstream processes to preserve temporal or linguistic context.
Compliance and Governance
Philter supports HIPAA Safe Harbor de-identification, aiding U.S. healthcare organizations in removing the 18 categories of identifiers from text-based records [2][5]. Since the tool relies on rules and dictionaries, the level of compliance depends on how thoroughly an organization configures and validates its filters against its specific clinical text [5]. For added assurance, organizations can use Expert Determination reviews, where a qualified expert evaluates re-identification risks and documents any remaining controls [5][8]. Additionally, healthcare organizations using broader risk management platforms, like Censinet RiskOps™, can document Philter's use as part of their privacy and cybersecurity framework. This allows them to track de-identification efforts alongside third-party risk assessments and vendor integrations involving clinical text [7].
Integration and Deployment
Philter is built for on-premises deployment, available as a command-line tool or containerized service for processing local files or streams [2][4][11]. This setup ensures that all data processing happens locally, which simplifies HIPAA compliance and Business Associate Agreement requirements by avoiding external APIs [4]. Many U.S. health systems incorporate Philter into ETL pipelines, nightly EHR exports, and data lakes. By placing it upstream of research warehouses and analytics platforms, they ensure only de-identified text reaches secondary systems [2][11]. Its BSD license and self-hosted model make it an attractive option for organizations seeking control over their infrastructure and predictable costs compared to cloud-based solutions.
3. NLM Scrubber
NLM Scrubber is a free and open-source tool developed by the U.S. National Library of Medicine to remove Protected Health Information (PHI) from unstructured clinical text. Unlike tools designed for images or structured databases, NLM Scrubber focuses specifically on narrative clinical text - the type of documentation commonly found in U.S. electronic health record (EHR) systems. Its rule-based design and ability to run locally make it an attractive option for academic institutions looking for a cost-free, secure de-identification solution. This targeted functionality makes NLM Scrubber a valuable addition to the broader ecosystem of de-identification tools.
Data Type Coverage
NLM Scrubber is tailored for free-text clinical documents, including discharge summaries, progress notes, operative reports, and pathology reports generated in U.S. healthcare settings [4]. By focusing on these text-heavy documents, the tool ensures that much of the clinical narrative remains intact for secondary use, while removing sensitive PHI. However, it does not support other data formats like structured HL7 or FHIR resources, DICOM imaging metadata, or database fields. Organizations working with these formats will need additional tools to handle those specific de-identification needs [2].
De-Identification Techniques
The tool uses a combination of rule-based pattern matching, curated dictionaries, and domain-specific heuristics to identify and redact PHI such as dates, phone numbers, and IDs [4][2]. Identified PHI is either removed or replaced with standardized placeholders, such as "[NAME]" or "[DATE]". For example, a patient's name might be replaced with "[NAME]" to ensure anonymity. Evaluations have reported a recall rate above 0.95 for detecting person names and IDs, though precision can occasionally suffer due to over-masking.
Organizations can further customize NLM Scrubber by adding site-specific data, such as local name lists, hospital names, and city names, to improve detection accuracy for their specific environment.
Compliance and Governance
NLM Scrubber is designed to meet regulatory requirements, making it a reliable choice for organizations operating under the stringent U.S. healthcare privacy framework. It supports both Safe Harbor and Expert Determination methodologies for de-identification. The tool enables HIPAA-compliant de-identification by systematically removing HIPAA's 18 identifiers from narrative clinical notes. For Safe Harbor compliance, it targets key identifiers such as names, geographic details smaller than a state, dates (except the year), phone numbers, and medical record numbers [5][8].
Under the Expert Determination method, privacy experts can use NLM Scrubber as a baseline tool, then assess residual risks through sampling, error analysis, or simulated attack scenarios [8][10]. Additionally, healthcare organizations using platforms like Censinet RiskOps™ can integrate NLM Scrubber into their broader privacy and cybersecurity frameworks, documenting its role in de-identification alongside other risk management efforts [7].
Integration and Deployment
Given the sensitivity of PHI, NLM Scrubber is typically deployed on-premises or within private cloud environments. It integrates into Extract, Transform, Load (ETL) workflows, processing notes extracted from EHR databases or data warehouses [4][2]. Many organizations run the tool as part of a nightly batch process, de-identifying new or updated notes before they are added to research repositories.
NLM Scrubber is distributed as a standalone application for Windows and Linux, with command-line configuration options. This setup makes it easy to integrate into workflows using Python, R, or shell scripts, as well as enterprise ETL tools or cron jobs [2]. By running locally, it eliminates the need for cloud dependencies or per-document API fees, though organizations must ensure proper resource allocation and monitoring to prevent any PHI exposure [7].
4. CliniDeID
CliniDeID builds upon existing tools to advance de-identification in clinical narrative processing. Developed by UTHealth (The University of Texas Health Science Center at Houston), this open-source de-identification system is tailored for removing PHI (Protected Health Information) from unstructured clinical narratives in U.S. healthcare environments [2]. Available on GitHub and supported by academic research, CliniDeID offers a cost-efficient alternative to commercial options, eliminating per-record or per-gigabyte fees [2][12]. Its transparency and customizable features make it a great choice for organizations that want full control over their de-identification rules. Like similar tools, it supports local processing and thorough validation to meet U.S. healthcare privacy regulations.
Data Type Coverage
CliniDeID is designed to handle unstructured clinical text found in EHRs (Electronic Health Records), such as discharge summaries, progress notes, consultation notes, radiology reports, and pathology reports [2]. These documents often contain a significant portion of PHI, making the tool especially useful for academic medical centers and research networks aiming to reuse clinical documentation for analytics or AI model development [2][5]. However, CliniDeID does not natively process DICOM imaging metadata, structured HL7 or FHIR resources, or highly structured database fields. Organizations working with these formats will need additional tools [2]. This targeted focus allows CliniDeID to excel in its specific niche.
De-Identification Techniques
CliniDeID employs a hybrid method that combines pattern-based rules (e.g., regular expressions) with lookup dictionaries. This approach identifies both direct and quasi-identifiers, such as patient and provider names, medical record numbers, account and encounter IDs, email addresses, street addresses, facility names, dates, ages, and other details closely aligned with HIPAA's 18 identifiers [2][3][8]. The tool can be configured to apply different transformations depending on the type of entity: full redaction for contact information, masking for partial phone or ID numbers, date shifting to maintain temporal relationships, or pseudonymization with consistent surrogates to support longitudinal studies while maintaining privacy [3][5]. This flexibility helps organizations meet various IRB or compliance requirements while balancing privacy and research needs [2][8].
Compliance and Governance
CliniDeID complies with both HIPAA Safe Harbor and Expert Determination standards, enabling the removal or transformation of all 18 HIPAA identifiers. It includes documented baselines, regular audits, and defined data access policies to monitor and mitigate re-identification risks [2][5][8]. For those using platforms like Censinet RiskOps™, CliniDeID can be integrated into broader privacy and cybersecurity systems, allowing for the tracking of de-identified datasets and periodic reassessment of re-identification risks as part of an organization’s overall risk management strategy [1][5][10].
Integration and Deployment
CliniDeID can be deployed as a batch de-identification step within ETL pipelines, typically on Linux servers or research clusters. It offers command-line and scriptable interfaces, making it easy to integrate into existing data workflows and NLP preprocessing pipelines [2][3]. In AI and analytics projects, CliniDeID is often used right before data exits a secure clinical environment, ensuring that narrative text is de-identified before being added to data lakes, model training systems, or shared collaboration platforms. Logs are maintained to verify consistent application of de-identification processes [2][3][7]. To ensure high-quality results, organizations can validate the tool’s performance through stratified sampling and iterative rule adjustments, while benchmarking throughput and latency to handle production data or large retrospective datasets [2][3][5][7][10].
5. ARX Data Anonymization
ARX Data Anonymization is an open-source tool built in Java that specializes in anonymizing structured, tabular datasets using formal privacy models. Unlike tools designed for handling unstructured clinical text or medical images, ARX shines when working with EHR extracts, patient registries, clinical trial datasets, and claims or billing tables. These datasets have clearly defined categories such as identifiers, quasi-identifiers, sensitive fields, and non-sensitive data [4]. Developed from academic research on privacy, ARX has been widely referenced in peer-reviewed studies and serves as a go-to solution for risk-based anonymization in healthcare, epidemiology, and clinical research. Its availability under the Apache 2.0 license makes it an appealing choice for U.S. health systems, academic institutions, and research groups operating on tight budgets [2]. ARX's focus on structured data sets it apart from tools designed for text or image de-identification.
Data Type Coverage
ARX is tailored for structured, tabular data formats like CSV files, relational database tables, and spreadsheet exports. In U.S. healthcare, this includes patient registries, outcomes research datasets, population health studies, and administrative claims files - datasets where rows typically represent patients or encounters, and columns contain demographic, clinical, or utilization attributes [4]. While ARX is highly effective for these data types, it does not handle unstructured text or medical images. For such data, organizations often use specialized de-identification tools first and then apply ARX as a second-stage anonymization tool for the resulting structured data. This targeted approach allows ARX to focus on delivering strong privacy protections for structured datasets while leaving other formats to more specialized tools.
De-Identification Techniques
ARX supports a variety of privacy models, including k-anonymity, ℓ-diversity, t-closeness, δ-presence, and β-likeness. It also offers differential privacy-based methods to anonymize numeric data [4]. To achieve these models, ARX employs techniques like:
- Generalization hierarchies: For instance, converting specific dates into broader categories like years or age ranges, or truncating ZIP codes to their first three digits.
- Suppression: Removing outlier records or rare attribute combinations.
- Microaggregation: Aggregating numeric fields into groups.
- Value-noising: Adding slight variations to preserve data distributions.
For example, a U.S. healthcare organization might use ARX to group dates of birth into age brackets (e.g., 0–17, 18–34, 35–49, 50–64, 65+), truncate ZIP codes to 3-digit prefixes, and aggregate ICD codes into broader categories. These techniques allow the organization to maintain analytical value while meeting k-anonymity thresholds [5][8]. ARX also provides robust risk metrics, such as prosecutor, journalist, and marketer risk models, along with average risk and equivalence class distributions. These metrics help balance privacy and data utility, ensuring datasets remain useful for clinical and operational analytics [4][5][8].
Compliance and Governance
ARX supports compliance with HIPAA Expert Determination and GDPR anonymization standards using its risk calculation framework and documentation features [13]. For Safe Harbor compliance, ARX can suppress or mask direct identifiers and generalize dates and locations to HIPAA-compliant levels (e.g., year-only for dates, 3-digit ZIP codes) [8]. For organizations pursuing Expert Determination, ARX provides tools for quasi-identifier modeling, probabilistic risk assessments, and detailed documentation of transformation rules. These features offer quantitative evidence to support an expert opinion that the "risk of re-identification is very small" [5][8][13].
To ensure comprehensive privacy protection, U.S. healthcare entities can integrate ARX into a formal data governance process. This might include documenting transformation rules, conducting expert reviews for high-risk data, performing periodic audits of ARX configurations, and implementing role-based access controls for both original and anonymized datasets [2][8]. Additionally, ARX outputs - such as de-identification methods, risk metrics, and data flow descriptions - can be integrated into platforms like Censinet RiskOps™ to enhance cybersecurity and risk management. This approach demonstrates a full-spectrum commitment to managing risks, from technical dataset controls to broader organizational governance.
Integration and Deployment
ARX is designed for seamless integration into ETL pipelines for structured data. It can be used as a stand-alone application with an intuitive GUI or integrated into automated ETL/ELT pipelines via APIs or scripting tools like Airflow or Databricks [6][7]. Common use cases include on-demand analysis for research requests or automated batch processing in data workflows. Performance depends on factors such as dataset size, the chosen privacy model (e.g., t-closeness can be computationally intensive), and the complexity of generalization hierarchies. For large datasets, such as those managed by U.S. health systems with millions of records, ARX is typically run on scaled compute nodes as part of batch processing, with teams monitoring execution times and risk metrics under service-level agreements [6][7]. Organizations often validate anonymization effectiveness and risk thresholds through iterative testing [5][8].
6. SDCmicro
SDCmicro is an open-source R package created by Statistics Austria for managing statistical disclosure control (SDC) in microdata. Unlike tools designed for handling unstructured data like text or images, SDCmicro focuses on structured, row-level data - think household surveys, census datasets, administrative records, or U.S. healthcare data such as electronic health record (EHR) extracts, patient registries, and claims files. It operates under the GPL-2 license, making it a cost-effective option for healthcare organizations, research institutions, and public health agencies that need reliable tools without breaking the bank. While ARX specializes in structured data anonymization through formal privacy models, SDCmicro excels at addressing the residual risks of re-identification in microdata.
Data Type Coverage
SDCmicro is specifically designed for microdata, where each row represents an individual record, and columns contain attributes like demographic details, clinical information, or service utilization. In the U.S. healthcare context, this includes datasets such as patient registries, hospital discharge data, population health studies, and administrative claims files. Its strength lies in managing residual re-identification risks tied to quasi-identifiers like age, ZIP code, gender, and diagnosis codes. However, it doesn't handle unstructured clinical text or medical images. To address this, organizations often start with tools that remove direct identifiers from text or images before using SDCmicro as a second-stage anonymization tool to handle structured data. This dual-step approach ensures SDCmicro can focus on structural de-identification in tabular data, complementing tools tailored for text or image anonymization.
De-Identification Techniques
SDCmicro offers a range of de-identification methods alongside tools for assessing risk. These include:
- Recoding (e.g., grouping ages into 5- or 10-year bands)
- Top- and bottom-coding for extreme values
- Local suppression of specific variables
- Microaggregation (clustering similar records and replacing values with cluster means or medians)
- Noise addition
- Rank swapping between records
- PRAM (post-randomization)
These techniques are particularly effective for U.S. healthcare datasets, significantly reducing re-identification risks while preserving the ability to calculate meaningful metrics like rates, odds ratios, and regression coefficients. SDCmicro also computes k-anonymity for each record, flagging those with low k-values (e.g., 1 or 2) as high-risk. Its documentation includes case studies showing how combining methods like recoding, suppression, and microaggregation can lower re-identification risks to acceptable levels while maintaining analytical accuracy within a few percentage points of the original data.
Compliance and Governance
SDCmicro helps quantify disclosure risks by identifying unique or rare records based on key variables, estimating re-identification probabilities, and assessing metrics like k-anonymity. Under HIPAA, while Safe Harbor mandates the removal of 18 specific identifiers, Expert Determination requires a documented analysis proving that re-identification risks are "very small." SDCmicro supports Expert Determination workflows by calculating metrics such as:
- Proportion of unique records in the sample
- Estimated uniqueness in the population
- Minimum and average k-values after transformations
- Maximum and average re-identification risks
Organizations can set internal thresholds, such as requiring a minimum k-value of 5 or 10 for high-risk combinations, and use SDCmicro's reports to confirm compliance. For health systems managing risks tied to third-party vendors, platforms like Censinet RiskOps™ can complement SDCmicro by centralizing vendor risk assessments and monitoring how third parties handle de-identified microdata.
Integration and Deployment
As an R package, SDCmicro integrates seamlessly into R-based ETL and analytics pipelines commonly used by research and healthcare analytics teams. U.S. organizations often deploy it in secure environments - either on-premises or in HIPAA-compliant cloud platforms - embedding it into scripted R workflows for consistent and reproducible risk analysis. Automated risk reports can be generated, and parameter settings (like age bands or geographic aggregation rules) can be logged in a configuration repository to ensure the process is both reproducible and auditable.
For those less familiar with programming, SDCmicro includes a GUI (sdcMicroGUI) for training and exploratory work. However, production workflows typically rely on scripted R code for reliability and automation. Teams using Python-heavy environments can also integrate SDCmicro by calling it via interop libraries or containerized R services, effectively wrapping it as a microservice that processes schemas and de-identification profiles. This approach fits well within broader ecosystems like Censinet RiskOps™, enabling organizations to manage both technical de-identification and vendor oversight in a unified framework.
sbb-itb-535baee
7. DICOMCleaner
When it comes to handling imaging data, DICOMCleaner stands out as a specialized tool for de-identifying medical images. Developed by PixelMed Publishing, this free, open-source, Java-based software is tailored to process medical imaging data in DICOM format. It's widely used across U.S. research institutions, academic medical centers, and multi-center imaging trials. DICOMCleaner ensures that datasets - spanning CT, MRI, X-ray, ultrasound, and more - are prepared for sharing, teaching, and AI development while safeguarding patient privacy.
Data Type Coverage
DICOMCleaner works with a variety of DICOM imaging data, including standard modalities such as CT, MR, CR, DX, US, PET, and secondary capture images. It can also process DICOMDIR media from CDs or DVDs [8]. The tool focuses primarily on metadata within DICOM headers, such as patient names, IDs, birth dates, accession numbers, study descriptions, and institutional details. Additionally, it can address burned-in identifiers (like text overlays in pixel data) by allowing users to crop or black out specific regions [4][1].
De-Identification Techniques
The tool applies standard DICOM confidentiality profiles to ensure sensitive data is protected. This includes suppressing, blanking, pseudonymizing, and date-shifting attributes [8]. Users can configure how tags are handled - whether retained, suppressed, or replaced - based on their organization's policies. DICOMCleaner also offers visual feedback, showing which tags are being modified, so privacy officers can review changes before finalizing them.
For added flexibility, the tool supports batch processing and can be operated interactively through its graphical interface or integrated into automated pipelines via command-line tools. In longitudinal studies, DICOMCleaner ensures consistent pseudonyms are applied to maintain a link between data sets without exposing PHI. This feature is particularly useful for research requiring patient data to remain identifiable across multiple exports.
Compliance and Governance
DICOMCleaner supports HIPAA-compliant de-identification under the Safe Harbor provision, allowing organizations to configure the removal of sensitive DICOM attributes and obscure burned-in text [5]. For scenarios requiring Expert Determination, a privacy expert can create custom de-identification profiles - such as partial date shifting or selective retention of non-identifying codes - and document that the risk of re-identification is minimal.
To enhance compliance, DICOMCleaner should be part of a broader governance framework that includes access controls, role-based permissions, and detailed logging of de-identification processes. Regular quality checks, whether manual or automated, are recommended to validate the de-identification process and ensure HIPAA requirements are met. For comprehensive risk management, the tool can also be integrated with larger cybersecurity platforms.
Integration and Deployment
As a Java-based, cross-platform tool, DICOMCleaner runs on Windows, macOS, and Linux. It can also be integrated into server-side or batch workflows using the PixelMed toolkit and command-line interface [6]. Typically, it serves as a gateway between production PACS/VNA systems and downstream systems for research, AI development, or teaching.
In practice, organizations extract studies from PACS, de-identify them using DICOMCleaner, and then route the sanitized data to research archives or cloud storage [6]. Many U.S. organizations deploy the tool on-premises for tighter control over PHI. However, some also use virtual machines in HIPAA-compliant cloud environments, with proper Business Associate Agreements, to handle larger datasets and integrate with cloud analytics platforms [7][9]. Combining DICOMCleaner with tools like NLP for text de-identification and an orchestration layer for data management creates a scalable solution for modern healthcare needs. This setup ensures efficient and secure handling of sensitive imaging data.
8. DICOM Anonymizer and PyDICOM with Deid
DICOM Anonymizer (a Windows-based GUI tool) and PyDICOM with Deid (a scriptable Python solution) are designed to remove identifying information from DICOM header metadata. These tools handle sensitive details like patient names, IDs, birth dates, and institution names across major imaging modalities such as CT, MR, XR, US, and PET. However, they do not address burned-in text within image pixels, so additional tools like OCR or vision-based solutions are necessary for complete removal of protected health information (PHI) [6][8].
Data Type Coverage
Both tools work with DICOM files from all major imaging modalities, including standard and many vendor-specific tags. Deid, in particular, uses YAML-based recipes to reference tags by keyword (e.g., PatientName) or full tag number, and it can incorporate the official DICOM data dictionary to handle custom fields. These tools primarily focus on metadata de-identification, ensuring identifiers in the header are suppressed, blanked, or replaced [6][8]. For multi-frame images, structured reports, or non-DICOM formats like JPEG, PNG, or NIfTI, additional conversion or processing may be needed. Manual rules and periodic audits are also recommended to address site-specific tags, especially after updates to PACS or imaging modalities.
De-Identification Techniques
Both tools rely on rule-based transformations aligned with HIPAA's 18 Safe Harbor identifiers. Common processes include:
- Suppression or removal: Blanking fields like patient name or address.
- Pseudonymization: Replacing identifiers with study-specific codes.
- Date adjustments: Shifting dates or generalizing them to retain only the year.
- Masking: Truncating details such as ZIP codes.
- Standardized replacements: Substituting fields like InstitutionName with "ANONYMIZED" [2][5][8].
DICOM Anonymizer uses a GUI with configurable tag lists, while PyDICOM with Deid employs YAML recipes for defining actions like remove, hash, date-shift, or replace. Deid also supports hashing and salting to irreversibly transform identifiers while maintaining consistency for longitudinal research. For example, a typical recipe might remove PatientName and PatientID, apply a random offset to StudyDate, and blank ReferringPhysicianName, with all changes logged for auditing [2][8]. These methods align with compliance protocols, ensuring technical de-identification integrates seamlessly into broader governance frameworks.
Compliance and Governance
For U.S. healthcare organizations, these tools support both HIPAA Safe Harbor and Expert Determination approaches. Safe Harbor configurations focus on removing or generalizing all 18 identifier categories in DICOM tags, prioritizing privacy through aggressive templates [2][5][8]. Expert Determination allows for more tailored templates that retain data utility - such as age ranges or broader geographic details - while requiring a documented risk assessment by a privacy expert [5][8].
Best practices include maintaining version-controlled templates (e.g., "safe_harbor.yml" for strict compliance vs. "expert_det_research.yml" for research needs), logging all de-identification activities, and conducting periodic reviews by privacy officers or radiologists. This structured approach aligns with modern de-identification frameworks, ensuring workflows integrate with broader governance and security programs. For data shared externally, organizations can incorporate findings into enterprise risk management tools like Censinet RiskOps™, which helps track cybersecurity and third-party risks throughout the data lifecycle.
Integration and Deployment
Deploying these tools efficiently balances PHI protection with operational needs. DICOM Anonymizer is typically used as a desktop GUI tool on Windows workstations, where users select DICOM studies, apply de-identification profiles, and export sanitized datasets to research archives or PACS. PyDICOM with Deid, on the other hand, is ideal for scripted workflows and integrates into pipelines for ETL tasks, AI training, or cloud-based processing.
Common deployment setups include on-premises services that read from PACS/VNA export directories, process files, and write de-identified outputs to research archives. In cloud environments, organizations often use storage buckets and containerized jobs triggered by REST APIs or message queues to run PyDICOM+Deid scripts. A 2021 audit of a hospital's imaging pipeline revealed that switching from manual DICOM editing to a PyDICOM-based automated workflow reduced processing time per study by 60–70%, enabling the team to handle tens of thousands of studies annually. For large-scale datasets, optimizing performance with batch processing, multiprocessing, and avoiding unnecessary pixel operations ensures efficient handling, while pilot testing helps validate workflows before full-scale deployment.
9. RSNA Image Share De-Identifier and Kheops De-Identification
The RSNA Image Share De-Identifier and Kheops De-Identification tools are designed to safeguard patient privacy within medical imaging workflows. The RSNA Image Share De-Identifier evolved from the RSNA Image Share project, which gave patients secure access to their imaging exams online. Kheops, created at the University Hospital of Geneva, is an open-source, web-based DICOM archive and sharing platform. It includes a built-in de-identification module specifically designed for research groups and collaborations across institutions. Together, these tools expand de-identification efforts from text and structured data to imaging workflows.
Data Type Coverage
Both tools focus on DICOM-based imaging workflows and accommodate all major imaging modalities. The RSNA Image Share De-Identifier removes or modifies protected health information (PHI) in DICOM headers and embedded pixel data, following HIPAA guidelines. Kheops De-Identification, on the other hand, is typically used in image-sharing and research repositories. It ensures uniform handling of DICOM headers, accession numbers, study identifiers, and linkage keys, so de-identified images remain searchable over time. While these tools primarily address imaging metadata, organizations often use separate natural language processing tools for associated radiology reports and HL7/FHIR messages.
De-Identification Techniques
The RSNA Image Share De-Identifier comes with DICOM de-identification profiles, such as "Basic" and "Retain Longitudinal with Modified Dates." These profiles allow for configurable date shifting and options to retain or remove specific device and institutional attributes, along with limited demographic details for research purposes. Kheops De-Identification emphasizes strong pseudonym generation and mapping tables, ensuring consistent patient and study linkage while enabling customization to preserve necessary modality information. Both tools remove high-risk PHI tags, apply date shifting, and pseudonymize identifiers, making longitudinal analysis possible without revealing actual identities. Administrators can customize tag-handling rules to meet institutional risk tolerances and IRB requirements, often blending Safe Harbor-style removal with selective retention under an Expert Determination framework.
Compliance and Governance
The RSNA Image Share De-Identifier includes predefined de-identification profiles that align closely with HIPAA Safe Harbor requirements. These profiles remove sensitive elements such as patient names, full-face photographs, device serial numbers, and detailed date information. Kheops, on the other hand, is often implemented under an Expert Determination framework. Privacy specialists document that its combination of tag removal, date shifting, and pseudonymization minimizes re-identification risks while maintaining research usability. Both tools are typically paired with strict governance measures, including version-controlled configurations and documented data-use agreements, to ensure compliance. Organizations may also integrate these workflows with platforms like Censinet RiskOps™ to standardize risk assessments for imaging PHI.
Integration and Deployment
These tools offer flexible deployment options tailored to imaging workflows. The RSNA Image Share De-Identifier is often positioned alongside existing PACS or VNA systems, acting as a gateway for de-identification before studies are exported to research archives, teaching files, or external collaborators. This on-premises setup supports horizontal scaling through multiple de-identification nodes, load-balanced routing, and asynchronous processing, ensuring clinical workflows remain uninterrupted. Kheops, by contrast, is commonly deployed in cloud-based imaging repositories for research and multi-institutional data sharing. It leverages DICOMweb, REST APIs, and token-based access to handle large-scale datasets efficiently, processing bulk cohorts at the archive level rather than on individual workstations. By 2025, many U.S. organizations are expected to adopt a hybrid model - using on-premises de-identification for operational exports and cloud-based systems like Kheops for large imaging datasets, AI development, and multi-center registries, all within HIPAA-compliant frameworks.
Strengths and Weaknesses
Choosing the right de-identification tool depends on factors like data types, workflows, and compliance requirements. The table below highlights how nine tools stack up across important dimensions for U.S. healthcare and research organizations.
| Tool | Primary Data Type | Key Strengths | Key Weaknesses | Best Fit |
|---|---|---|---|---|
| PhysioNet DeID | Clinical text (free-text notes) | Open-source, research-tested, and uses a transparent rule-based system that's easier for IRBs to audit | Struggles with nuanced identifiers compared to modern NLP tools, limited support for newer formats like FHIR, and requires technical expertise for dictionary tuning | Academic centers working with PhysioNet datasets seeking reproducible and auditable methods |
| Philter | Clinical text (plain text notes) | Excels at identifying direct identifiers (names, dates, IDs); allows custom blacklists and batch processing for large datasets | Focuses mainly on direct identifiers, leaving potential gaps with quasi-identifiers; lacks a robust GUI and built-in reporting for Expert Determination documentation | Research environments needing batch de-identification of notes in HIPAA-regulated, on-premises setups |
| NLM Scrubber | Clinical text (narrative notes) | Developed by the National Library of Medicine; uses transparent NLP methods; addresses all 18 HIPAA identifiers; ideal for reproducible IRB protocols | Limited ongoing development compared to commercial tools and requires expertise for tuning and EHR integration | Research institutions needing government-supported, reproducible methods for clinical narratives |
| CliniDeID | Clinical text (clinical narratives) | Offers customizable rulesets and dictionaries tailored to institutional PHI patterns | Demands significant technical expertise for tuning and assessing residual re-identification risks; lacks enterprise-ready features out of the box | Organizations with in-house data science teams requiring flexible text de-identification solutions |
| ARX Data Anonymization | Structured/tabular data (EHR extracts, registries, claims) | Provides k-anonymity, l-diversity, and t-closeness; features a visual interface for balancing risk and utility; well-suited for Expert Determination workflows | Steep learning curve for non-statisticians; lacks native support for unstructured text or images; requires ETL engineering | Health systems releasing structured datasets under Expert Determination while focusing on preserving data utility |
| SDCmicro | Structured/tabular data (microdata, registries) | Strong statistical disclosure control tools with transparent, reproducible methods and granular risk metrics, widely used in official statistics | Requires proficiency in R; limited integration with clinical data pipelines; less healthcare-specific documentation for HIPAA Safe Harbor mapping | Research teams with statistical expertise managing structured dataset releases |
| DICOMCleaner | DICOM imaging | Handles large imaging archives efficiently; offers standard tag removal and a user-friendly GUI; supports typical research export workflows | Limited to DICOM data; users must configure tag retention to balance utility and HIPAA compliance | Radiology departments exporting imaging studies for research or educational purposes |
| DICOM Anonymizer / PyDICOM with Deid | DICOM imaging | Highly flexible with programmatic control for automated pipelines; supports pixel data redaction, such as facial blurring in CT/MRI | Requires significant technical expertise and rigorous testing; lacks built-in compliance documentation; places governance responsibility on configuration | Advanced research teams integrating de-identification into automated PACS-to-research or cloud storage workflows |
| RSNA Image Share De-Identifier / Kheops | DICOM imaging | Designed to align with radiology standards; validated for multi-institutional image sharing; supports research repositories | Limited scope (imaging only, not enterprise-wide PHI); requires integration into broader compliance frameworks; DICOM header de-identification alone may not suffice without pixel data control | Multi-center imaging research networks and radiology collaborations needing standardized workflows |
When choosing a tool, compliance and governance are just as important as functionality. Tools that align with HIPAA Safe Harbor make compliance easier but can reduce the utility of research data. On the other hand, tools like ARX and SDCmicro offer advanced anonymization models and re-identification risk metrics, making them better suited for Expert Determination workflows. However, it's crucial to remember that no tool can guarantee complete de-identification. Studies have shown that even de-identified clinical notes can be vulnerable to attacks like membership inference, underscoring the need for expert legal and statistical review [10].
Costs and support models also influence tool selection. For U.S. health systems managing PHI risks across various platforms, integrating de-identification tools with broader cybersecurity frameworks like Censinet RiskOps™ can streamline risk assessments and documentation.
Ultimately, the trade-off is clear: stricter de-identification reduces risk but can limit data utility, while more flexible methods require robust governance. Testing tools on your institution's data is essential before full deployment, as vendor-reported accuracy often varies across different datasets, whether they are clinical notes, imaging studies, or structured data [5]. Platforms like Censinet RiskOps™ can assist in this process by helping healthcare organizations document de-identification workflows alongside enterprise risk management efforts.
Conclusion
When selecting de-identification tools, it’s crucial to choose options that align with your specific data type and workflow. Many U.S. healthcare organizations rely on a mix of specialized solutions to address their unique needs. This approach reflects the diverse strategies discussed earlier.
To make an informed choice, focus on practical selection criteria such as the deployment model, local accuracy validation, the impact on data utility, and the total cost of ownership. Piloting multiple tools on actual clinical data is essential. During this process, monitor how well each tool detects PHI and use human quality reviews to identify any residual errors.
These recommendations build on the detailed benchmark analyses shared earlier. As Matt Christensen, Sr. Director GRC at Intermountain Health, aptly states, "Healthcare is the most complex industry... You can't just take a tool and apply it to healthcare if it wasn't built specifically for healthcare." To enhance your strategy, integrate de-identification workflows with broader cybersecurity platforms like Censinet RiskOps™. This integration helps document vendor risk assessments, maintain audit trails, and ensure alignment with enterprise PHI protection practices.
Striking the right balance between effective de-identification and preserving data utility requires strong governance and continuous expert validation. Before full deployment, test tools on your datasets, and standardize de-identification processes. Clear policies, role-based access controls, and integration with risk management platforms are critical to making de-identification a seamless preprocessing step in your organization.
FAQs
How do de-identification tools help meet regulations like HIPAA and GDPR?
De-identification tools play a crucial role in helping organizations comply with regulations like HIPAA and GDPR. They use methods such as data masking, anonymization, and pseudonymization to safeguard sensitive information, ensuring that personal data - like protected health information (PHI) - can’t be linked back to individuals.
To support compliance, these tools often come equipped with features like audit trails, role-based access controls, and real-time monitoring. These capabilities help document and secure how data is handled. Additionally, they are built to keep up with evolving legal standards, giving organizations the resources they need to manage and protect sensitive data securely.
How do de-identification tools differ for clinical text, structured data, and medical imaging?
De-identification tools are designed to handle specific types of data, tailoring their methods to suit the format and requirements of each. When it comes to clinical text, these tools rely on natural language processing (NLP) and AI to locate and mask protected health information (PHI) in free-text notes. The goal is to maintain the usefulness and meaning of the data while ensuring sensitive details are hidden.
For structured data, such as databases or spreadsheets, the tools focus on anonymizing patient identifiers. This is often achieved through rule-based systems or machine learning algorithms that prioritize precision and compliance with privacy standards.
In the case of medical imaging, advanced image processing techniques are employed. These methods obscure identifiable features, like facial details in scans, all while preserving the diagnostic quality of the images to ensure they remain clinically valuable.
The main distinction between these tools lies in the type of data they handle - whether it's text, structured formats, or images - and the unique techniques required to process each securely and effectively.
How can healthcare organizations ensure privacy while keeping data useful with de-identification tools?
Healthcare organizations can safeguard patient privacy while still making data useful by using specialized de-identification tools. These tools are designed to reduce the risk of re-identifying individuals and ensure compliance with privacy regulations, all while protecting sensitive information like patient details and Protected Health Information (PHI).
With these purpose-designed solutions, organizations can securely share and analyze data for clinical and research needs. This approach allows them to maintain compliance and operational efficiency without sacrificing the utility of the data.


